 |
精彩产品 |
 |
|
|
|
|
|
 |
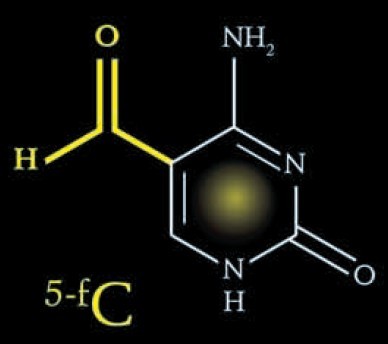
| 精彩产品>表观遗传学..热销.. |
RNA甲基化修饰和定量?惊呆了我和小伙伴们!!!--new |
 |
 |
 |
 |
| 组蛋白H3多通道修饰分析试剂盒(比色法) (A-P-3100) |
| Input materials can be histone extracts or purified histone H3 proteins. The amount of histone extracts for each assay can be 20 ng to 500 ng with an optimal range of 50 ng to 100 ng depending on the purity of histone extracts. The amount of purified hist
|
|
 |
|
|
 |
 |
 |
 |
| 组蛋白H4多通道修饰分析试剂盒(比色法) (A-P-3102) |
| Input materials can be histone extracts or purified histone H4 proteins obtained from human, mouse, rat, as well as a broad range of species including most plants, fungi, and bacteria, based on high sequence homology of histone H4. The amount of histone e
|
|
 |
|
|
 |
 |
 |
 |
| 高敏免疫沉淀试剂盒 (A-P-2027) |
| Starting materials can include various tissue or cell samples such as cells from flask or plate cultured cells, fresh and frozen tissues, etc. In general, the amount of cells and tissues for each reaction can be 2 x 103 to 1 x 106 and 0.5 mg to 50 mg, res
|
|
 |
|
|
 |
 |
 |
 |
| 高敏免疫测序试剂盒 (A-P-2030) |
Starting materials can include various tissue or cell samples such as culture cells from a flask or plate, fresh and frozen tissues, etc.
|
|
 |
|
|
 |
 |
 |
 |
| 现货---EZ RNA甲基化修饰试剂盒 (A-R5001) |
Fast and reliable bisulfite conversion of RNA for methylation analysis.
Specifically optimized for complete conversion of non-methylated cytosine in RNA.
Ideal for all RNA inputs.
|
|
 |
|
|
|
|
 |
 |
 |
 |
| 亚硫酸盐快速DNA文库制备试剂盒(illumina) (A-P-1055) |
| Input starting material must be bisulfite-treated DNA generated from various input DNA amounts of 0.5 ng to 1 µg. For optimal preparation, the input DNA amount for the bisulfite conversion process should be 100 ng to 200 ng so that sufficient bisulfite-tr
|
|
 |
|
|
 |
 |
 |
 |
| 高敏DNA文库制备试剂盒(illumina) (A-P-1053) |
| Starting materials can include fragmented dsDNA isolated from various tissue or cell samples, dsDNA enriched from a ChIP reaction, MeDIP/hMeDIP reaction, or exon capture. DNA should be relatively free of RNA because large fractions of RNA will impair end
|
|
 |
|
|
 |
 |
 |
 |
| DNA文库制备试剂盒(illumina) (A-P-1051) |
| Starting materials can include fragmented dsDNA isolated from various tissue or cell samples, dsDNA enriched from ChIP reactions, MeDIP/hMeDIP reaction, or exon capture. DNA should be relatively free of RNA since large fractions of RNA will impair end rep
|
|
 |
|
|
|
| |
|
|
|
| |
|